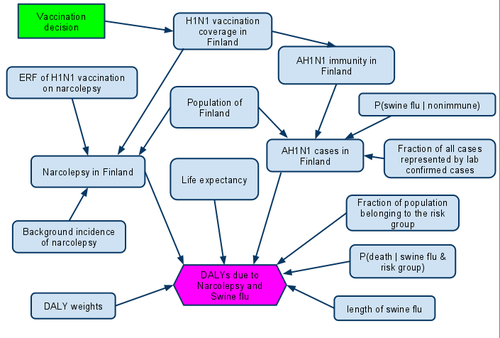
+ Show code- Hide code
# Model; original data inputs are disability weights (isfw, inarcw), population (ipop/dpop), (atm) ERF of vaccine on narcolepsy (NERF),
# vaccination coverage (ivac_cov), base immunity in the population (ibimm) and observed number of sf cases (dsf)
# Function for aggregating indicators into DALYs
# Narcolepsy DALY weight taken as 0.065 (that of treated epilepsy)
# iEl is expectation of remaining life in an age group, value for "All" is arbitrary for now
# ilsf is the length of swine flu (e.g. how long was the subject absent frow work) in years
# ifrg is the fraction of population belonging to a risk group of dying from swine flu, default values are arbitrary
# iElrgc is the expected life correction for the risk group (people with heart conditions etc are likely to die earlier)
outcome <- function(inarc = narc(), isf = sf(), inarcw = data.frame(Result=0.065),#op_baseGetData("opasnet_base", "Op_en2307")[,-c(1,2)],
isfw = data.frame(Result=0.5), iEl = data.frame(Age=c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49",
"50-54","55-59","60-64","65-69","70-74","75-79","80+","All"), Result = c(79,75.1,70.1,65.1,60.3,55.5,50.7,45.9,41.1,36.5,31.9,27.6,
23.5,19.5,15.7,12.1,9,50)), ilsf = data.frame(Result=5/365), isfpd = sfpd(), ifrg = data.frame(Age = c("0-4 ","5-9","10-14","15-19",
"20-24","25-29","30-34","35-39","40-44","45-49","50-54","55-59","60-64","65-69","70-74","75-79","80+"), Result = c(0.1, rep(0.001,
11), rep(0.5,5))), iElrgc = data.frame(Age = c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49","50-54",
"55-59","60-64","65-69","70-74","75-79","80+"), Result = c(rep(1,12), rep(0.5,5))), ...) {
names(inarc)[names(inarc)=="Result"] <- "inarc"
names(inarcw)[names(inarcw)=="Result"] <- "inarcw"
names(isf)[names(isf)=="Result"] <- "isf"
names(isfw)[names(isfw)=="Result"] <- "isfw"
names(iEl)[names(iEl)=="Result"] <- "iEl"
names(ilsf)[names(ilsf)=="Result"] <- "ilsf"
names(isfpd)[names(isfpd)=="Result"] <- "isfpd"
names(ifrg)[names(ifrg)=="Result"] <- "ifrg"
names(iElrgc)[names(iElrgc)=="Result"] <- "iElrgc"
# Calculating narcolepsy DALYs
onarc <- merge(inarc, inarcw)
onarc <- merge(onarc, iEl)
onarc <- model.frame(I(inarc * inarcw * iEl) ~., data = onarc)
colnames(onarc)[1] <- "onarc"
# Calculating swine flu DALYs from missing work
osf <- merge(isf, isfw)
osf <- merge(osf, ilsf)
osf <- model.frame(I(isf * isfw * ilsf) ~., data = osf)
colnames(osf)[1] <- "osf"
#Calculating swine flu DALYs from deaths
osfm <- merge(isf, isfpd)
osfm <- merge(osfm, iEl)
osfm <- merge(osfm, ifrg)
osfm <- merge(osfm, iElrgc)
osfm <- model.frame(I(isf * isfpd * iEl * ifrg * iElrgc) ~., data = osfm)
colnames(osfm)[1] <- "osfm"
# Aggregate (it is crucial that both variables are distinguished by the same age groups and other indices so that values
# are not replicated to fill empty cells)
out <- merge(onarc, osf)
out <- merge(out, osfm)
out <- reshape(out, idvar = names(out)[(names(out)%in%c("onarc","osf","osfm"))==FALSE], times = c("onarc","osf","osfm"),
timevar = "Outcome", varying = list(c("onarc","osf","osfm")), direction = "long")
colnames(out)[ncol(out)] <- "Result"
rownames(out) <- 1:nrow(out)
return(out)
}
# Function for calculating narcolepsy cases, depends on erf of vac on narc, pop, vac_cov and background incidence (fraction)
narc <- function(iNERF = op_baseGetData("opasnet_base", "Op_en4923")[,-c(1)], ipop = op_baseGetData("opasnet_base", "Op_en2949")[,-c(1,2)],
ivac_cov = op_baseGetData("opasnet_base", "Op_en4925")[,-c(1,2)], ibgi = data.frame(Result=1.4e-5), ...) {
names(iNERF)[names(iNERF)=="Result"] <- "iNERF"
names(ipop)[names(ipop)=="Result"] <- "ipop"
names(ivac_cov)[names(ivac_cov)=="Result"] <- "ivac_cov"
names(ibgi)[names(ibgi)=="Result"] <- "ibgi"
out <- merge(iNERF, ipop)
out <- merge(out, ivac_cov)
out <- merge(out, ibgi)
out <- model.frame(I((iNERF - 1) * ipop * ivac_cov * ibgi) ~., data = out)
colnames(out)[1] <- "Result"
return(out)
}
# A function for calculating iNERF might be added later
# Function for calculating swine flu cases
# ilcf is the fraction of actual swine flu cases represented by the lab confirmed cases
# assumed binomially distributed, if uncertainty of probability of getting swine flu is not
# present, the point estimate will be used as such and n samples sampled from the same distribution;
# however if the probability has an uncertainty, only 1 sample from each distribution is sampled where
# samples of the probability will be used as inputs; disabled sampling here since it would make different scenarios incomparable
sf <- function(isfp = sfp(), ipop = op_baseGetData("opasnet_base", "Op_en2949")[,-c(1,2)], iimm = imm(), n = 1000,
ilcf = tlcf, ...) {
names(isfp)[names(isfp)=="Result"] <- "isfp"
names(ipop)[names(ipop)=="Result"] <- "ipop"
names(iimm)[names(iimm)=="Result"] <- "iimm"
names(ilcf)[names(ilcf)=="Result"] <- "ilcf"
out <- merge(isfp, ipop)
out <- merge(out, iimm)
out <- merge(out, ilcf)
#Result <- NA
#for (i in 1:nrow(out)) {
# if (sum(as.numeric(colnames(isfp)=="obs"))>0){
# Result[i] <- rbinom(1, round(out$ipop[i]*(1-out$iimm[i])), out$isfp[i]) / out$ilcf[i]
# } else {
# Result[((i-1)*n + 1):(i*n)] <- rbinom(n, round(out$ipop[i]*(1-out$iimm[i])), out$isfp[i]) / out$ilcf[i]
# }
#}
#out <- data.frame(out[,(colnames(out)%in%c("isfp","ipop","iimm","ilcf"))==FALSE], Result)
out <- model.frame(I(isfp * ipop * (1 - iimm) / ilcf) ~., data = out)
colnames(out)[1] <- "Result"
return(out)
}
# Function for calculating probability of getting swine flu given subject is not vaccinated (simplified model)
# dsf, dpop and dimm are the actual data we have
# uncertainty: number of cases is binomially distributed, number of trials (n) is the number of people who were not immune to swine-flu
# outcome (x) is known, p is unknown. P can be easily calculated from x = p*n, when we assume that x is the expected value of the
# distribution. But we're interested in the uncertainty of the estimate... When n is suitably large a poisson distribution becomes
# an excellent approximation of the binomial distribution. So we can fit the poisson distribution using an R function (I didn't find
# one for binomial) from just one observation -> acquire value of x with standard error -> use as uncertainty of estimate; which produces
# rubbish because we only have a single observation, sd of the mean estimate of the poisson distribution appears to be (n*p)^0.5, so the
# variance of the parameter is n * p, which divided by n gives p ... so the expected value is p and the variance is p, which causes problems
# with the beta distribution (shape parameters become negative), hence variance is assumed to be variance^2 arbitrarily
sfp <- function(dsf = op_baseGetData("opasnet_base", "Op_en4933")[,-c(1,2)], dpop = op_baseGetData("opasnet_base", "Op_en2949",
series_id = 970)[,-c(1,2)], dimm = imm(), n = 1000, ...) {
names(dsf)[names(dsf)=="Result"] <- "dsf"
names(dpop)[names(dpop)=="Result"] <- "dpop"
names(dimm)[names(dimm)=="Result"] <- "dimm"
temp1 <- merge(dimm, dpop)
temp1 <- model.frame(I(dpop * (1 - dimm)) ~., data = temp1)
colnames(temp1)[1] <- "popnonimm"
out <- merge(temp1, dsf)
temp2 <- data.frame(obs=NA, Age=NA, fitp=NA)
out2 <- data.frame()
for (i in 1:nrow(out)) {
temp2 <- data.frame(obs=1:n, Age=out$Age[i], fitp=rbeta(n, paralpha(fitdistr(round(out$dsf[i]), "poisson")[[1]] /
out$popnonimm[i], (fitdistr(round(out$dsf[i]), "poisson")[[2]]^2 / out$popnonimm[i])^2),
parbeta(fitdistr(round(out$dsf[i]), "poisson")[[1]] / out$popnonimm[i], (fitdistr(round(out$dsf[i]),
"poisson")[[2]]^2 / out$popnonimm[i])^2)))
out2 <- rbind(out2, merge(out[i,], temp2, all.x = TRUE, all.y = FALSE))
}
out2 <- out2[,(colnames(out2)%in%c("dsf", "popnonimm"))==FALSE]
colnames(out2)[ncol(out2)] <- "Result"
return(out2)
}
# Function for calculating probability of mortality after catching swine flu given that a subject belongs to a risk group
# children of age < 1 and adults of 60+ afflicted by heart conditions etc are considered to belong to a risk group
# ifrg is the fraction belonging to the risk group in a given age group
# p(death|sf&rg)) assumed constant in all age groups
sfpd <- function(dsf = op_baseGetData("opasnet_base", "Op_en4933")[,-c(1,2)], dmsf = data.frame(Result=44),
dfrg = data.frame(Age = c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49",
"50-54","55-59","60-64","65-69","70-74","75-79","80+"), Result = c(0.1, rep(0.001,11), rep(0.5,5))), ilcf = tlcf, ...) {
names(dsf)[names(dsf)=="Result"] <- "dsf"
names(dmsf)[names(dmsf)=="Result"] <- "dmsf"
names(dfrg)[names(dfrg)=="Result"] <- "dfrg"
names(ilcf)[names(ilcf)=="Result"] <- "ilcf"
out <- merge(dsf, dfrg)
out <- merge(out, ilcf)
#out <- merge(out, dmsf)
out <- model.frame(I(dfrg * dsf / ilcf) ~., data = out)
colnames(out)[1] <- "Result"
if (sum(as.numeric(colnames(out)=="obs"))>0) {#out <- as.data.frame(as.table(tapply(out$)))
out <- data.frame(obs = levels(factor(out$obs)), Result = dmsf$dmsf/tapply(out$Result, out$obs, sum))
} else {out <- data.frame(Result=dmsf$dmsf/sum(out$Freq))}
return(out)
}
# Function for calculating total immunity to swine flu in the whole population
# P(imm) = P(vac or base_imm)
imm <- function(ibimm = op_baseGetData("opasnet_base", "Op_en4943", series_id = 994)[,-c(1,2)],
ivac_cov = op_baseGetData("opasnet_base", "Op_en4925")[,-c(1,2)], ...) {
names(ibimm)[names(ibimm)=="Result"] <- "ibimm"
names(ivac_cov)[names(ivac_cov)=="Result"] <- "ivac_cov"
out <- merge(ibimm, ivac_cov)
out <- model.frame(I(1 - (1 - ibimm) * (1 - ivac_cov)) ~., data = out)
colnames(out)[1] <- "Result"
return(out)
}
# Calculating the outcome with different scenarios
paralpha <- function(imean, ivar) imean * (imean * (1 - imean) / ivar - 1)
parbeta <- function(imean, ivar) (1 - imean) * (imean * (1 - imean) / ivar - 1)
library(OpasnetBaseUtils)
library(MASS)
library(ggplot2)
# Common variable generation
n <- 1000
tlcf <- data.frame(obs = 1:n, Result=rbeta(n, paralpha(0.2, 0.1^2), parbeta(0.2, 0.1^2)))
#ggplot(tlcf, aes(x=Result)) + geom_density()
#tlcf <- data.frame(Result = 0.1)
tsfp <- sfp()
#test2 <- data.frame(obs = 1:n, Result=rbeta(n, paralpha(0.2, 0.2^2+0.01), parbeta(0.2, 0.2^2+0.01)))
#ggplot(test2, aes(x=Result)) + geom_density()
# Default, model based on current knowledge
# fast version below
#vacscenario <- outcome(inarc = narc(iNERF = data.frame(Age = c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49",
# "50-54","55-59","60-64","65-69","70-74","75-79","80+","All"), Result = c(1,rep(9.2, 3), rep(1, 13), 4))))
#novacscenario <- outcome(inarc = narc(iNERF = data.frame(Age = c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49",
# "50-54","55-59","60-64","65-69","70-74","75-79","80+","All"), Result = c(1,rep(9.2, 3), rep(1, 13), 4)),
# ivac_cov=data.frame(Result=0)), sf(iimm=imm(ivac_cov=data.frame(Result=0))))
#colnames(vacscenario)[ncol(vacscenario)] <- "vacscenario"
#colnames(novacscenario)[ncol(novacscenario)] <- "novacscenario"
#fout <- merge(vacscenario, novacscenario)
#fout
# slow ver below
tvac_cov <- data.frame(op_baseGetData("opasnet_base", "Op_en4925")[,-c(1,2)], Scenario = "vacscenario")
tvac_cov <- rbind(tvac_cov, data.frame(Age=tvac_cov[,"Age"], Result = 0, Scenario = "novacscenario"))
test <- outcome(inarc = narc(ivac_cov = tvac_cov), sf(isfp = tsfp, iimm = imm(ivac_cov = tvac_cov)))
temp <- as.data.frame(as.table(tapply(test$Result, test[,c("obs","Scenario")], sum)))
ggplot(temp, aes(x = Freq, fill = Scenario)) + geom_density(alpha = 0.2)
tapply(temp$Freq, temp[,c("Scenario")], mean)
#op_baseWrite("opasnet_base", test)
# Sensitivity and value of information analysis
#test <- op_baseGetData("opasnet_base", "Op_en4926")
temp2 <- as.data.frame(as.table(tapply(test$Result, test[,(colnames(test)%in%c("Result","Outcome"))==FALSE], sum)))
tNERF <- op_baseGetData("opasnet_base", "Op_en4923")[,-c(1)]
colnames(tNERF)[colnames(tNERF)=="Result"] <- "tNERF"
colnames(tlcf)[colnames(tlcf)=="Result"] <- "tlcf"#tlcf from above
colnames(tsfp)[colnames(tsfp)=="Result"] <- "tsfp"#tsfp from above
temp2 <- merge(temp2, tNERF)
temp2 <- merge(temp2, tlcf)
temp2 <- merge(temp2, tsfp)
temp2$NERFbin <- cut(temp2$tNERF, c(-Inf, 1, quantile(temp2$tNERF[temp2$tNERF>1], 1:9/10), Inf))
temp2$lcfbin <- cut(temp2$tlcf, c(-Inf, quantile(temp2$tlcf, 1:9/10), Inf))
temp2$sfpbin <- cut(temp2$tsfp, c(-Inf, quantile(temp2$tsfp, 1:9/10), Inf))
temp3 <- as.data.frame(as.table(tapply(temp2$Freq, temp2[,(colnames(temp2)%in%c("Freq","obs","tNERF","tlcf","tsfp",
"lcfbin","sfpbin"))==FALSE], mean)))
colnames(temp3)[ncol(temp3)] <- "EogNERF"
temp2 <- merge(temp2, temp3)
temp3 <- as.data.frame(as.table(tapply(temp2$Freq, temp2[,(colnames(temp2)%in%c("Freq","obs","tNERF","tlcf","tsfp",
"NERFbin","sfpbin","EogNERF"))==FALSE], mean)))
colnames(temp3)[ncol(temp3)] <- "Eoglcf"
temp2 <- merge(temp2, temp3)
temp3 <- as.data.frame(as.table(tapply(temp2$Freq, temp2[,(colnames(temp2)%in%c("Freq","obs","tNERF","tlcf","tsfp",
"lcfbin","NERFbin","EogNERF","Eoglcf"))==FALSE], mean)))
colnames(temp3)[ncol(temp3)] <- "Eogsfp"
temp2 <- merge(temp2, temp3)
## Correlation
cor(temp2$Freq, temp2[,c("tNERF", "tlcf", "tsfp")], method = "spearman")
## EVPI
mean(tapply(temp$Freq, temp$obs, min)) - min(tapply(temp$Freq, temp$Scenario, mean)) # not considering
## EVXPI for NERF, lcf and sfp
mean(apply(tapply(temp2$EogNERF, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin",
"EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - min(tapply(temp$Freq, temp[,c("Scenario")], mean))
mean(apply(tapply(temp2$Eoglcf, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin",
"EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - min(tapply(temp$Freq, temp[,c("Scenario")], mean))
mean(apply(tapply(temp2$Eogsfp, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin",
"EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - min(tapply(temp$Freq, temp[,c("Scenario")], mean))
#vacscenario <- outcome()
#novacscenario <- outcome(inarc = narc(ivac_cov=data.frame(Result=0)), sf(iimm=imm(ivac_cov=data.frame(Result=0))))
#temp <- as.data.frame(as.table(tapply(vacscenario$Result, vacscenario[,c("obs","Outcome")], sum)))
#tapply(vacscenario$Result, vacscenario[,c("Age","Outcome")], mean)
#tapply(temp$Freq, temp$Outcome, mean)
#ggplot(temp, aes(x=Freq, fill=Outcome)) + geom_density(alpha = 0.2)
#colnames(vacscenario)[ncol(vacscenario)] <- "vacscenario"
#colnames(novacscenario)[ncol(novacscenario)] <- "novacscenario"
#fout <- merge(vacscenario, novacscenario)
#fout <- reshape(fout, idvar = names(fout)[(names(fout)%in%c("vacscenario","novacscenario"))==FALSE], times = c("vacscenario","novacscenario"),
# timevar = "Scenario", varying = list(c("vacscenario","novacscenario")), direction = "long")
#colnames(fout)[ncol(fout)] <- "Result"
#temp <- as.data.frame(as.table(tapply(fout$Result, fout[,c("obs","Scenario")], sum)))
#ggplot(temp, aes(x = Freq, fill = Scenario)) + geom_density(alpha = 0.2)
# What actually happened
#defscenario <- outcome(inarc = op_baseGetData("opasnet_base", "Op_en4922")[,-1], isf = op_baseGetData("opasnet_base", "Op_en4933")[,-1])
# Narcolepsy erf = 1
#nerfscenario <- outcome(iNERF = data.frame(Result=1))
# No one was vaccinated
#novacscenario <- outcome(ivac_cov = 0)
# Other
| |